|
Test-of-Time Award
|
|
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 
AbstractPredicting patient features from single-cell data can unveil cellular states implicated in health and disease. Linear models and average cell type expressions are typically favored for this task for their efficiency and robustness, but they overlook the rich cell heterogeneity inherent in single-cell data. To address this gap, we introduce GMIL, a framework integrating Generalized Linear Mixed Models (GLMM) and Multiple Instance Learning (MIL), upholding the advantages of linear models while modeling cell-state heterogeneity. By leveraging predefined cell embeddings, GMIL enhances computational efficiency and aligns with recent advancements in single-cell representation learning. Our empirical results reveal that GMIL outperforms existing MIL models in single-cell datasets, uncovering new associations and elucidating biological mechanisms across different domains. |
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 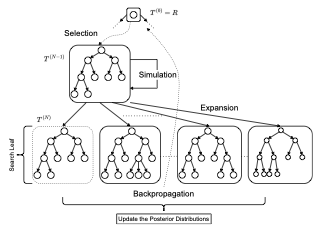
AbstractDecision Trees are prominent prediction models for interpretable Machine Learning. They have been thoroughly researched, mostly in the batch setting with a fixed labelled dataset, leading to popular algorithms such as C4.5, ID3 and CART. Unfortunately, these methods are of heuristic nature, they rely on greedy splits offering no guarantees of global optimality and often leading to unnecessarily complex and hard-to-interpret Decision Trees. Recent breakthroughs addressed this suboptimality issue in the batch setting, but no such work has considered the online setting with data arriving in a stream. To this end, we devise a new Monte Carlo Tree Search algorithm, Thompson Sampling Decision Trees (TSDT), able to produce optimal Decision Trees in an online setting. We analyse our algorithm and prove its almost sure convergence to the optimal tree. Furthermore, we conduct extensive experiments to validate our findings empirically. The proposed TSDT outperforms existing algorithms on several benchmarks, all while presenting the practical advantage of being tailored to the online setting. |
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 
AbstractAccess to pre-trained models has recently emerged as a standard across numerous machine learning domains. Unfortunately, access to the original data the models were trained on may not equally be granted. This makes it tremendously challenging to fine-tune, compress models, adapt continually, or to do any other type of data-driven update. We posit that original data access may however not be required. Specifically, we propose Contrastive Abductive Knowledge Extraction (CAKE), a model-agnostic knowledge distillation procedure that mimics deep classifiers without access to the original data. To this end, CAKE generates pairs of noisy synthetic samples and diffuses them contrastively toward a model’s decision boundary. We empirically corroborate CAKE's effectiveness using several benchmark datasets and various architectural choices, paving the way for broad application. |
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 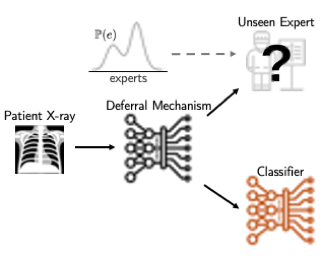
AbstractThe learning to defer (L2D) framework allows autonomous systems to be safe and robust by allocating difficult decisions to a human expert. All existing work on L2D assumes that each expert is well-identified, and if any expert were to change, the system should be re-trained. In this work, we alleviate this constraint, formulating an L2D system that can cope with never-before-seen experts at test-time. We accomplish this by using a meta-learning architecture for the deferral function: given a small context set to identify the currently available expert, the model can quickly adapt its deferral policy. We also employ an attention mechanism that is able to look for points in the context set that are similar to a given test point, leading to an even more precise assessment of the expert's abilities. In the experiments, we demonstrate the usefulness of this architecture on image recognition, traffic sign detection, and skin lesion diagnosis benchmarks. |
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 
AbstractWe propose Functional Flow Matching (FFM), a function-space generative model that generalizes the recently-introduced Flow Matching model to operate directly in infinite-dimensional spaces. Our approach works by first defining a path of probability measures that interpolates between a fixed Gaussian measure and the data distribution, followed by learning a vector field on the underlying space of functions that generates this path of measures. Our method does not rely on likelihoods or simulations, making it well-suited to the function space setting. We provide both a theoretical framework for building such models and an empirical evaluation of our techniques. We demonstrate through experiments on synthetic and real-world benchmarks that our proposed FFM method outperforms several recently proposed function-space generative models. |
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 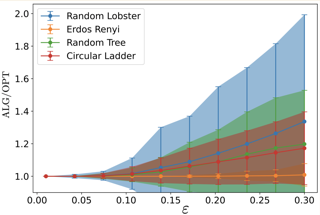
AbstractWe consider the problem of graph searching with prediction recently introduced by Banerjee et al. (2023). In this problem, an agent starting at some vertex r has to traverse a (potentially unknown) graph G to find a hidden goal node g while minimizing the total distance traveled. We study a setting in which at any node v, the agent receives a noisy estimate of the distance from v to g. We design algorithms for this search task on unknown graphs. We establish the first formal guarantees on unknown weighted graphs and provide lower bounds showing that the algorithms we propose have optimal or nearly-optimal dependence on the prediction error. Further, we perform numerical experiments demonstrating that in addition to being robust to adversarial error, our algorithms perform well in typical instances in which the error is stochastic. Finally, we provide simpler performance bounds on the algorithms of Banerjee et al. (2023) for the case of searching on a known graph and establish new lower bounds for this setting. |
|
Student Paper Highlight
|
Poster
[ MR1 & MR2 ] 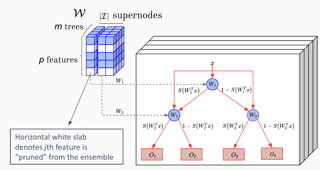
AbstractJoint feature selection and tree ensemble learning is a challenging task. Popular tree ensemble toolkits e.g., Gradient Boosted Trees and Random Forests support feature selection post-training based on feature importances, which are known to be misleading, and can significantly hurt performance. We propose Skinny Trees: a toolkit for feature selection in tree ensembles, such that feature selection and tree ensemble learning occurs simultaneously. It is based on anend-to-end optimization approach that considers feature selection in differentiable trees with Group L0 − L2 regularization. We optimize with a first-order proximal method and present convergence guarantees of our algorithmic approach for a non-convex and non-smooth objective. Interestingly, dense-to-sparse regularization scheduling can leadto more expressive and sparser tree ensembles than vanilla proximal method. On 15 synthetic and real-world datasets, Skinny Trees can achieve 1.5x− 620x feature compression rates, leading up to 10× faster inference over dense trees, without any loss in performance. Skinny Trees lead to superior feature selection than many existing toolkits e.g., in terms of AUC performance for 25% feature budget, Skinny Trees outperforms LightGBM by 10.2% (up to 37.7%), and Random Forests by 3% (up to 12.5%). |